Material linked from bioinformatics course
Group 1: Data
- What are the major strengths and the major weaknesses of the SwissProt database?
- What are the major strengths and the major weaknesses of the PDB database?
- What are the differences between SwissProt and UNIPROT?
- What is the role of the accession code in sequence databases?
- What are the major advantages and disadvantages of WWW based servers?
- What is the EBI?
- Mention five types of industrie that make intensive use of biomolecular data and databases,
and mention briefly how they use those facilities.
- If you obtain a novel sequence, which database searches do you perform first?
- Which are the five data types that every good database minimally should provide?
- What data does the EMBL database provide regarding the translation from DNA to protein?
- What kind of data is stored in (O)MIM.
- What is (O)MIM an abbreviation of?
- What is the difference between a SNP and a mutation mentioned in (O)MIM?
|
Video with the answers for group 1.
|
Group 2: Software
- What kind of searches can be done with MRS?
- What kind of searches can be done with BLAST?
- Why don"t we teach students how to operate many programs?
- What is CLUSTAL?
- Design a question for a bioinformatics examination, the correct answering of which
requires knowledge/usage of BLAST, MRS and CLUSTAL.
- During the course we only searched in SwissProt. That was for efficiency reasons. What
should you do in real research? Give advantages and disadvantages of your aproach in just
a few words.
|
Video with the answers for group 2.
|
Group 3: The amino acids
- Which residues are hydrophobic?
- Which residues are hydrophilic?
- What is special about proline?
- What have Thr, Ile and Val in common?
- What is special about glycine?
- What is special about cysteine?
- What is special about histidine?
- Why are Gly, Asp, Asn and Ser more likely in β-turns?
- Why are Ile, Val and Thr good residues for a β-strand?
- Write down the names of the large residues.
- Which are the three smallest residues?
- Which two residues cause different turns and bends of the backbone from all others?
- Mention two types of mutations (i.e. Phe -> Asp or Trp -> Ala) that generally make
proteins more stable.
- Why is Leucine not a good active site residue?
- Why is Cysteine seldomly seen at the surface of a protein?
- Why is Proline often observed at the surface of a protein?
- Why is Methionine often observed at the surface of a protein?
- Why is Tryptophan the most conserved residue?
- Why is it important to know all these characteristics of amino acids?

|
Video with the answers for group 3.
|
Group 4: Secondary structure
- What is the difference between a β-strand and a β-sheet?
- What is helix capping?
- Which residues are good for a helix?
- Which residues are good for a strand?
- Which residues are found most often in β-turns?
- At which end of a helix can you find prolines? And why?
- Why is the capping effect by charged amino acids stronger at the N-terminal end than at
the C-terminal end of a helix?
- How many amino acids are there in one helical turn?
- Briefly describe the hydrogen bonding pattern in a helix.
|
Video with the answers for group 4.
|
Group 5: Secondary structure prediction
- Which is the better helix A or B?
A: SPAALKALAEAAGS
B: SGAALKALAEAAPS
- Which is the better helix A or B?
A: PAAKALAEAASS
B: PAAEALAKAASS
- Which is the better strand A or B?
A: STVVTTIITTGS
B: STFKVTIRITGS
- Which is the better strand A or B?
A: SLATELLLAEKS
B: SIATLEIAIETS
- Predict the secondary structure of TVAPGTIE
- Predict the secondary structure of TIEVKITS
- Predict the secondary structure of PAELALKA
- Predict the secondary structure of SANTPRGS
- Predict the secondary structure of EEEEEEEE
- Give the sequence of the "perfect" helix (use about 10 amino acids, all different)
- Give the sequence of the "perfect" strand (use about 10 amino acids, all different)
- Give the sequence of the "perfect" β-hairpin (use about 10 amino acids, all different)

|
Video with the answers for group 5.
|
Group 6: Protein structure
- What is meant with primary-, secondary-, tertiary- and quarternary structure of proteins?
- What is entropy and what is enthalpy?
- Explain each of the terms in δG=δH-TδS in no more than 10 words each.
- What is the difference between a protein and an enzyme?
- Why doesn't there exist an enzyme with an acive site consisting of Leu, Val and Phe?
|
Video with the answers for group 6.
|
Group 7: Sequence alignment
- Why is sequence alignment such an important tool?
- What is the relation between sequence alignment and evolution?
- What is a gap penalty?
- What is a Dayhoff matrix?
- A Dayhoff matrix is symmetric. Why is that a problem?
- What are the major considerations when designing a Dayhoff-like matrix?
- Align without gaps:
LLLDAWLLL with LLLDWLLL
- Align without gaps:
LLLPATLLL with LLLPTLLL
- Align without gaps:
LLLCARLLL with LLLCRLLL
- Align:
NRTSPEQLAL with NGGPDLAEVL
- Align:
ALALAKVPDV with LLKAGGTDWR
- Align:
KFTMGNMMWE with IRCPSFTMRR
|
Video with the answers for group 7.
|
Group 8: Other questions:
- What is PROSITE?
- Your newly discovered sequence contains the PROSITE pattern [S,T],{P},[K,R],{P} What should
you do before you publish this pattern?
- When do you use PAM30 and when PAM200 in BLAST?
- When do you use Blosum45 and when Blosum80 in BLAST?
- What does "Filter low-complexity regions:" mean in BLAST.
- When should you use the low complexity filter in BLAST and when not?
- What do we mean with a forced marriage?
- Draw a sequence of 14 amino acids of which the middle ten are perfectly helical and the outer
two at both ends not.
- Why do we need to learn how to align sequences by hand while computer programs exist that can
do it for you?
|
Video with the answers for group 8.
|
Group 9: Other questions:
- What was the main lesson you learned in this bioinformatics course?
Practical part of exam
The exam sometimes holds a small experimental part that must be solved using the
computer. If the group gets bigger than 30 students this option becomes impossible...
The experimental parts always are open-book. In 2008 we asked two questions
(after 2008 we never did a practical part again):
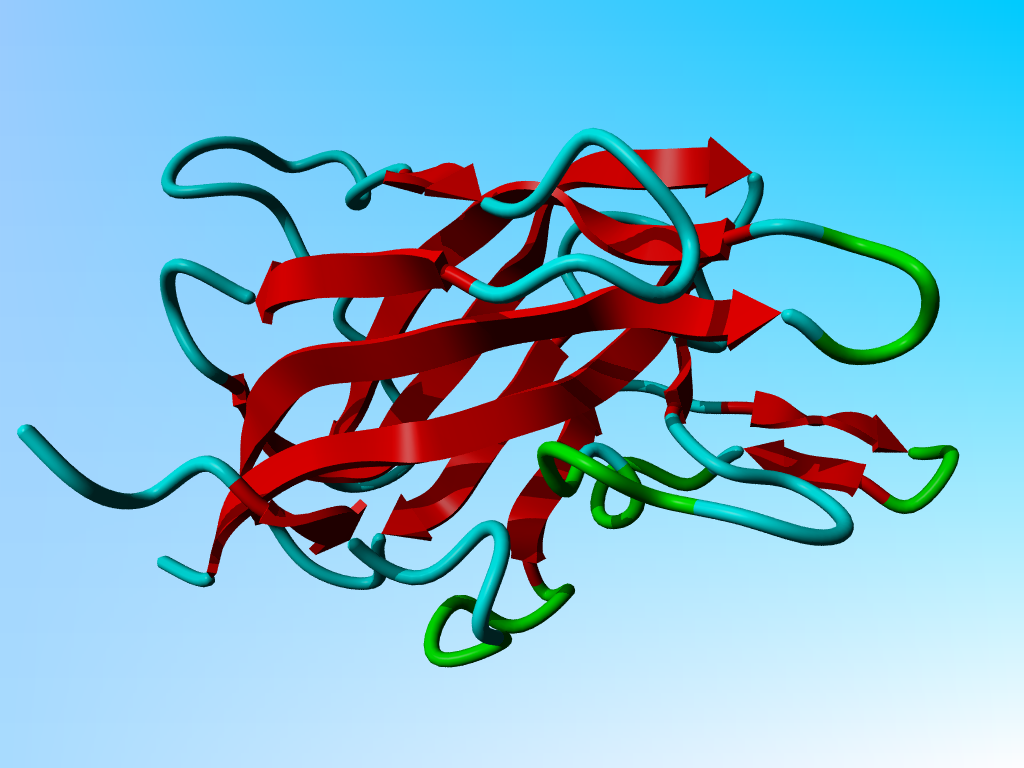
Coagulation factor 5A
|
The structure of the bovine factor 5A coagulation co-factor has been solved
by X-ray, and the structure has been deposited in the PDB with identifier
1SDD. Please think where you store this file! Don't forget the rules...
|
In the exercise files section
you will find 1SDD_clean.PDB. That PDB file holds just one
of the two (highly similar) domains that this protein uses to dock on a
membrane (so it is not a transmembrane protein).
Question 1
The questions is how this protein docks to the membrane. In other words, which
side of the protein touches the membrane and how does it do that.
There are two rather different ways of approaching this problem.
The one involves Google; the other involves YASARA. We want you to do both.
Look at the molecule with YASARA and find how this molecule can anchor itself
(and a whole series of attached domains that I have removed for you) to a
membrane. Explain briefly how the relation between residue characteristics and
function helped you find the answer. With Google you should also be able
to find a good site (preferably one that agrees with your own answer...).
Write down the WWW-address of the page you found (please write it down
carefully, I might need to type it in to check things) and copy one key
sentence that tells the whole story from the WWW page, or direct me to the
crucial picture on that page.
The answer
Look at the structure and you see two very exposed Tryptophans next to each other. Those two
would, of course, love to be in a membrane. Just next to them there are several positive residues that
love to interact with the phosphate head-groups of the lipid membranes. The easiest way to find this
idea confirmed is to MRS for the whole 1SDD file. Get the article out (The crystal structure of activated
protein C-inactivated bovine factor Va: Implications for cofactor
TE Adams, MF Hockin, KG Mann, SJ Everse - Proceedings of the National Academy of Sciences, 2004 -
National Acad Sciences). Find it with Google Scholar, or get it directly from the PNAS site, and look
for figure 4.
Question 2
Shortly after giving several students grades ranging from 1+ till 3- a bioinformatics professor was
found dead behind his terminal. The NCSI crime scene investigators found that he was killed with a
South-American poisoned arrow. They still found enough of the poison on the arrow that stuck in the
professors neck to determine its sequence: LACAPRGGILCYRCKECCKGLTCKGKFVNTWPTRCVV
To estimate the time of death, the investigators wanted to look at the decay of the poison in the victims
body. This decay is characterized by the reaction of one of the cysteines in this poison with a
cysteine-rich blood hormone that we all normally have in our circulation. Which is the Cysteine in
the poison that the NCSI crime scene investigators should concentrate on?
The answer
Take the sequence. Run Blast against swissprot. You get about nine hits. Activate them all and send them
to Clustal. Add our sequence before running as our sequence is not in the database yet. Run clustal.
All cysteines are conserved except for one in our sequence, the third C, that is the one between the R
and the K. To be sure you can check the cysteine bridge annotation in some of the aligned SwissProt files.
Other solution. Run Blast against the PDB. You find 1 or 2 hits. Load one structure in YASARA.
1LRM is an NMR structure so you must click some things away before getting a clean view. You now see
the cysteine bridges. Find these three pairs of cysteines back in the Blast alignment. The one not aligned
is the free one.
(Note added many years later: more sequences entered the database over the years. Nowadays (2015)
Clustal does not align all cysteines perfectly any longer (newer sequences are too different to align as
well as the easy ones), but it is still clear, albeit not exam-clear, that the third Cys does not occur
in the other sequences).








