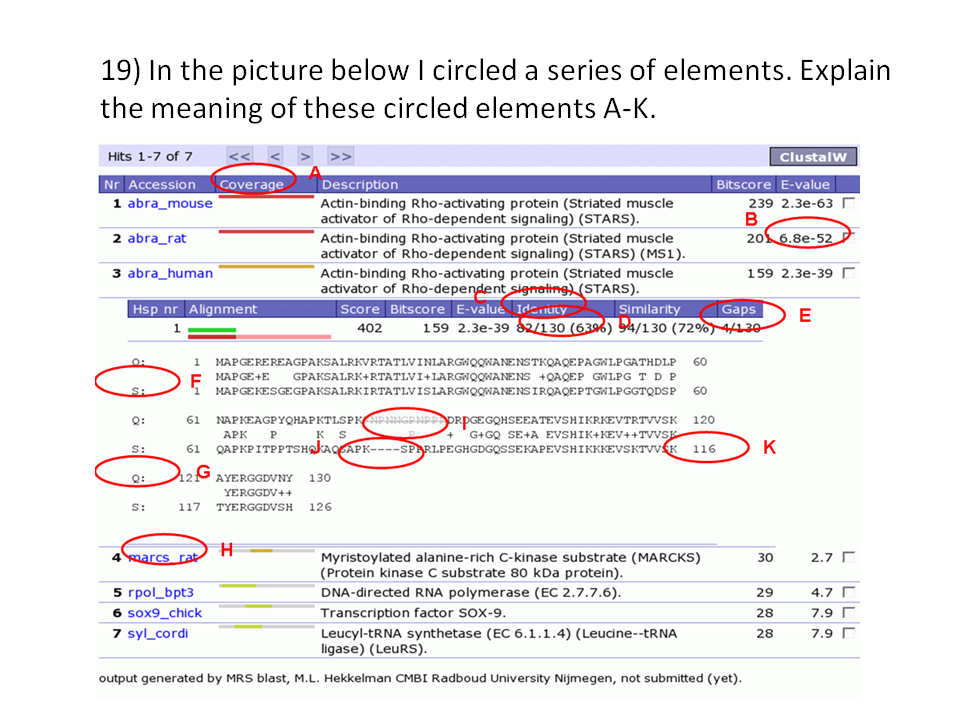
Annotated BLAST output.

|
Annotated BLAST output. |
This is NOT the BLAST output of CRAM_CRAAB, but the output of a somewhat longer sequence
the BLAST output of which contains all possible aspects of the output format.
A) The coloured coverage bar indicates which parts of something align with something.
B) De E-value indicates the quality of the alignment (how many hits would you find
by chance alone). Above 10-6 things gets dodgy.
C/D) C is the title of the identity field, and D holds the actual identity percentage between
your query sequence and the database subject sequence.
E) As C and D together, but now for the number of gaps in the alignment.
F/G) Q indicates your query sequence and S indicates the database subject sequence.
H) SwissProt file name of the 4-th hit sequence.
I) The short area that is scratched out is a low-complexity area. Other BLAST servers
put X-es there.
J) The dashes indicate a gap in the alignment.
K) The numbers before and after the sequence bars are the sequence numbers of the first
and the last residue on that line.
Further, some on the non-circled areas:
Similarity is a score that is better not used because it is subjective whether one
calls two residues similar or not. So, multiple BLAST servers can happily give the same hit with the
same percentage identity but a different percentage similarity.
The Score is simply all scores for the aligned residues added up. Using some complicated math this gets
converted into a Bit Score that is normalized in such a way that they can be compared between
runs. And a second step of math converts this into the E-value.