The NewProt efforts will all revolve around the protein engineering self-service portal, called SSP. The SSP will hold a large series of software, database, and information facilities. Table 1, to the left, lists a low-detail summary; details are provided in WP2-5. The SSP will be used for most dissemination activities, for the distribution of software and databases, and for all NewProt management activities. The SSP will include interactive workbench facilities. All facilities will become fully interoperable which will allow for easy to use yet powerful predictive and analytic protein engineering possibilities. The design of the SSP will get the highest priority in the first 12 months of the project.

The software and database partners will in the first 12 months prepare a comprehensive set of their own protein engineering software products for installation at or coupling to the SSP. They will work on the curation of commonly used, freely available databases and prepare these for installation (installation implies full integration and interoperability).
The experimental validation partners will in the first 12 months select a series of protein engineering (and chemical biology) targets that they will use for validation of the predictive capabilities of the available software packages, and they will set-up everything needed for engineering these proteins.
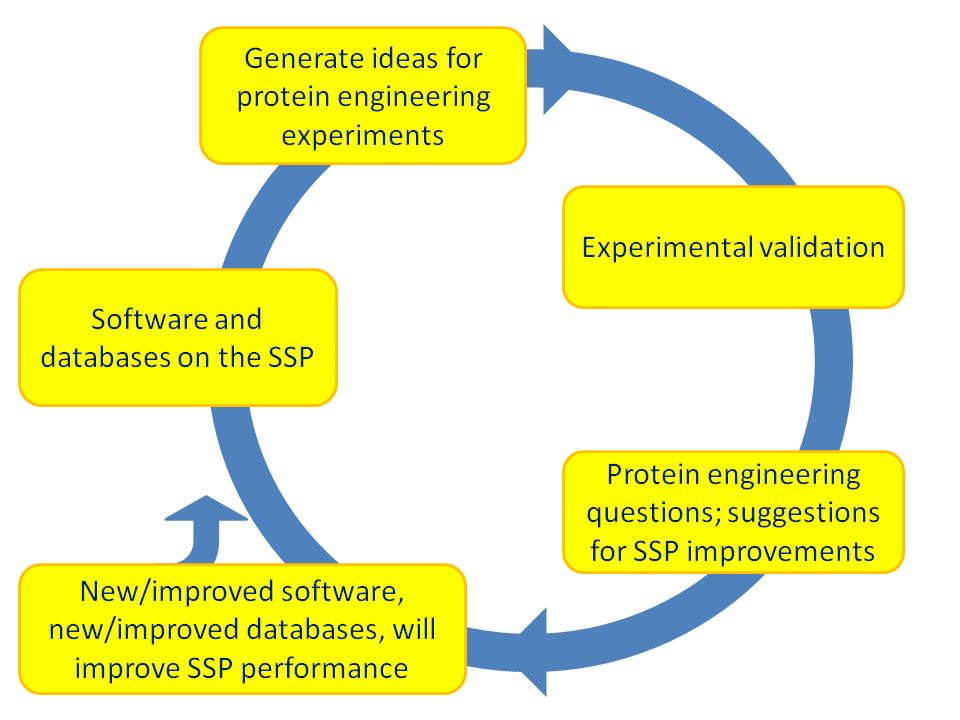
After the initial 12 months the partners will run the iterative regime shown in figure 1.2.
 |
Figure 1.2. The iterative cycle of protein engineering. The cycle can start at each position: Software generates ideas that can be experimentally validated, while lab work generates questions that can be answered with software. The cycle will lead to improved software, improved protocols, improved proteins, and thus to improved science in general. |
Like most first generation bioinformatics software, much protein engineering software suffers from an interface designed for specialist leading to a steep learning curve. The NewProt partners will make sure that all software will be easy to use, will have good explanations and help-facilities, and that supervisor systems will provide aid for the optimal use of those software products. The latter will be especially important for small research groups and SMEs that cannot afford a large bioinformatics department that can solve computer software usage problems for them.
In summary, the NewProt partners will design the first comprehensive protein engineering self-service portal, called SSP, that will be of benefit to all researchers in this field, but most to those who do not have a large in-house bioinformatics department. The SSP will be solidly based on modern portal and work-flow technology to allow for flexible usage, maintenance, distribution, and dissemination.