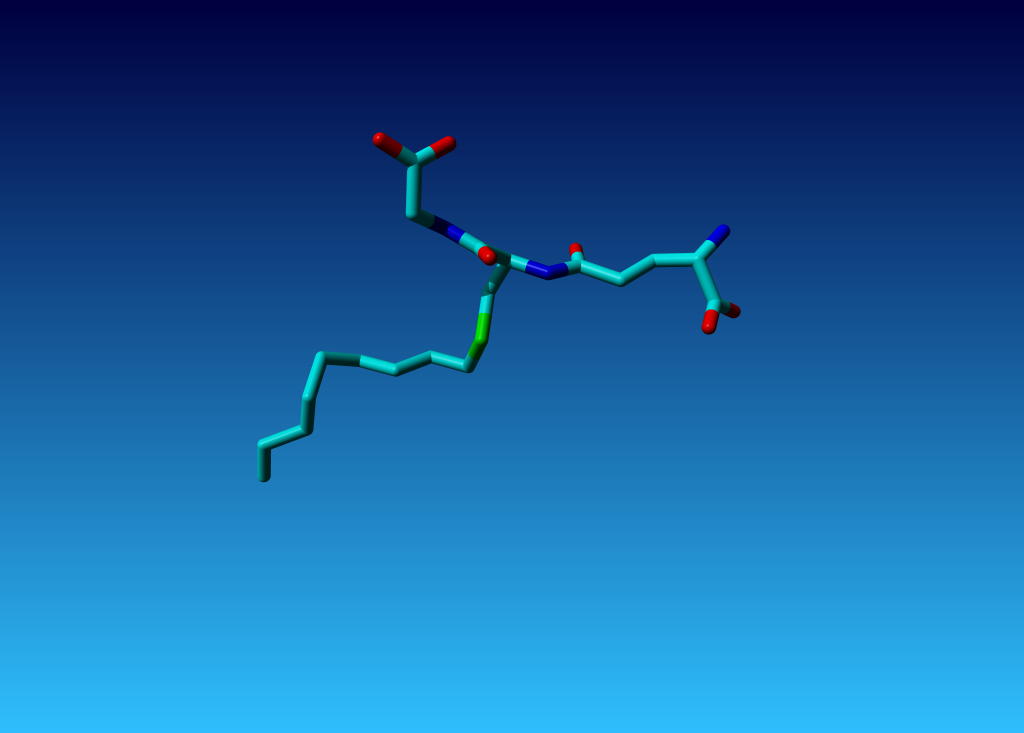
Figure 7. The funny tripeptide. The residues are coloured in the next picture for easy reference in this picture.
Often ligands are made of amino acids and 'funny things'. Green fluorecent protein is a famous example that makes its own prostetic group out of three amino acids. In 12GS we find something funny too:
JRNL AUTH A.J.OAKLEY,M.LO BELLO,M.NUCCETELLI,A.P.MAZZETTI, JRNL AUTH 2 M.W.PARKER JRNL TITL THE LIGANDIN (NON-SUBSTRATE) BINDING SITE OF HUMAN JRNL TITL 2 PI CLASS GLUTATHIONE TRANSFERASE IS LOCATED IN THE JRNL TITL 3 ELECTROPHILE BINDING SITE (H-SITE). JRNL REF J.MOL.BIOL. V. 291 913 1999 |
This PDB file contains two copies of a funny ligand that consists of three 'amino acids':
SEQRES 1 C 3 GLU GT9 GLY SEQRES 1 D 3 GLU GT9 GLY MODRES 12GS GT9 C 2 CYS S-NONYL-CYSTEINE MODRES 12GS GT9 D 2 CYS S-NONYL-CYSTEINE |
And in these funny tripeptides the glutamic acid is bound to the backbone N of the modified cysteine via a dehydrolysis involving its sidechain Oε2. We actually don't have the tools yet to check that it really is the Oε2 and not the Oε1...

|
Figure 7. The funny tripeptide. The residues are coloured in the next picture for easy reference in this picture. |
|
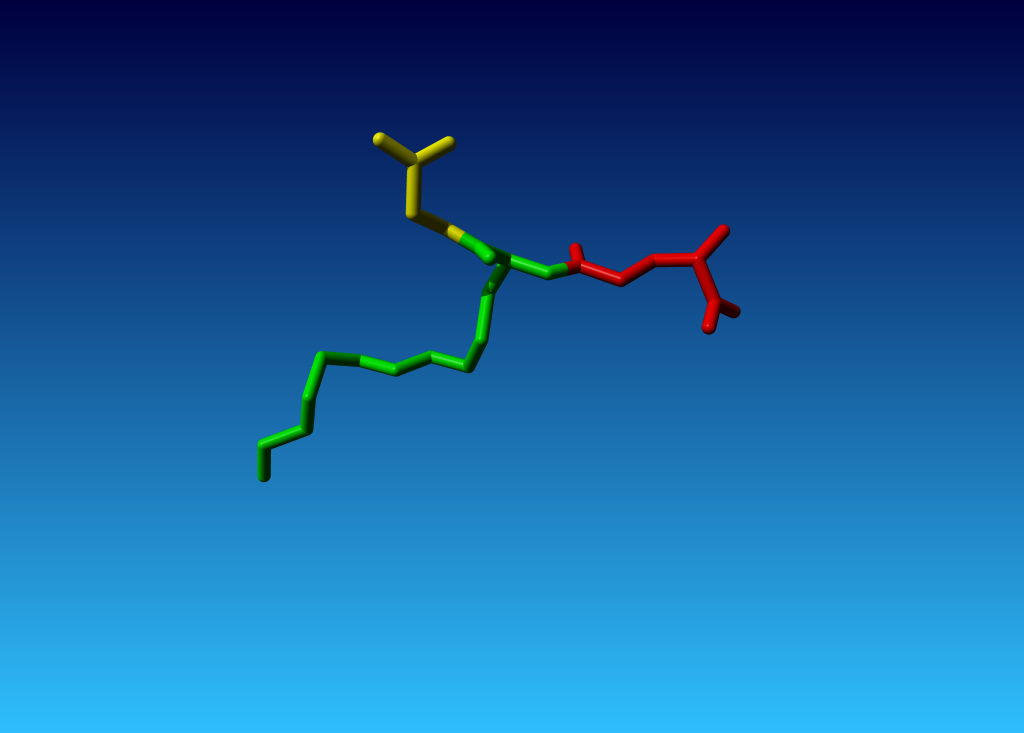
Figure 8. Same tripeptide as in the previous picture. Glu is red; modified Cys in green. Gly in yellow. |
Of course there is nothing wrong with this ligand, but it is very inconvenient for many bioinformatics applications that this thing has Glu and Gly in it. Why not simply make it one molecule named LIG or so? Much software will make mistakes with these residues in ligands.