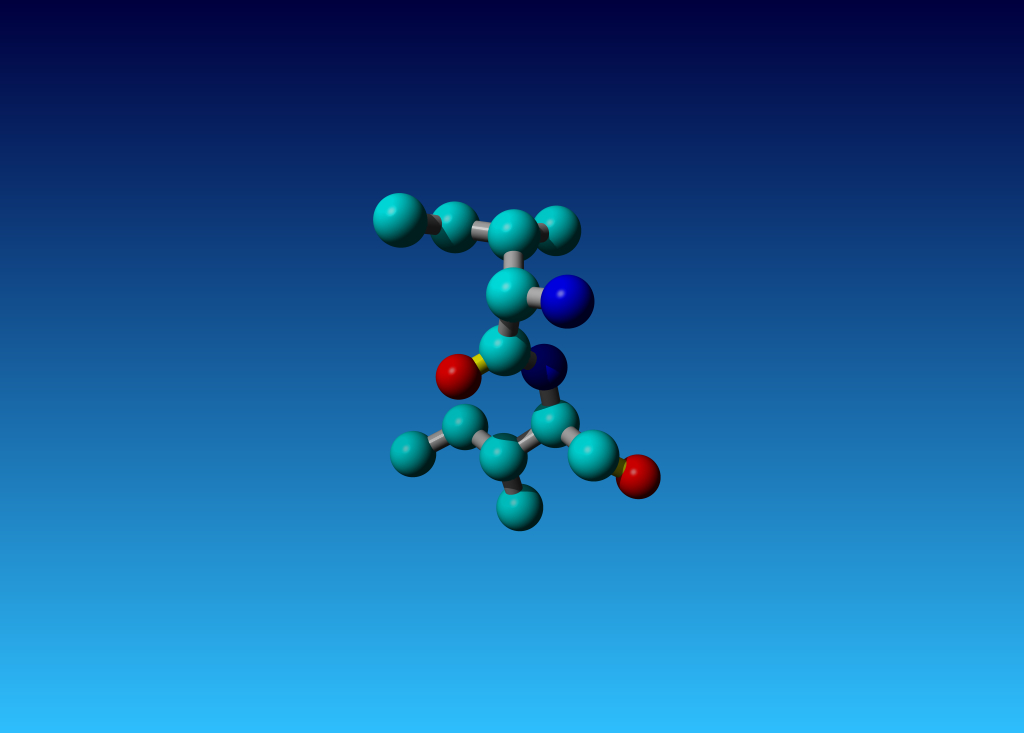
Figure 35. Isoleucines 10 and 11 in MODEL 2 of 1ACP.
Some aspects of chirality have been mentioned already in the section with administrative problems. Here I will elaborate a bit more on this topic.
There are a few genuine d-amino acids in the PDB. For example gramicidin a. But I think that in most cases this is an error. I cannot give you much advice for what to do when you run into this problem. As a user of the structure, I guess you should read if the d-amino acids are real or not, and if they are erroneous, look for another structure. And if you are a crystallographer or an NMR spectroscopist, get help from a senior, start fresh using a homology model of your structure, or Email me for help.
A more hidden problem is the chirality on the Cβ of isoleucine. And I present one example here of a really bad case.
1ACP is a very old NMR structure. But as it was used to test the use of molecular dynamics in NMR structure refinement, I was a bit surprised to find many chirality problems. In the note below you find the abstract of the article describing 1ACP.
Supplemental materialIn 1ACP I found these two isoleucines next to each other. They are residues 10 and 11 in the second MODEL.

|
Figure 35. Isoleucines 10 and 11 in MODEL 2 of 1ACP. |
It was just funny to see that Isoleucine 10 is OK (in terms of chirality) while residue 11 has the wrong chirality on both the Cα and the Cβ.
WHAT_CHECK tells us about 1ACP:
Three numbers are given for each atom in the table. The first is the Z-score for the improper dihedral. The second number is the measured improper dihedral. The third number is the expected value for this atom type. A final column contains an extra warning if the chirality for an atom is opposite to the expected value. 7 VAL ( 7-) A 1 CA -43.4 -30.19 33.23 Wrong hand 8 LYS ( 8-) A 1 C -7.2 -10.86 0.10 10 ILE ( 10-) A 1 CA -42.0 -31.81 33.24 Wrong hand 11 ILE ( 11-) A 1 CA -42.8 -33.10 33.24 Wrong hand 29 VAL ( 29-) A 1 CA -45.3 -32.85 33.23 Wrong hand 46 LEU ( 46-) A 1 CA -45.4 -35.25 34.17 Wrong hand 85 LYS ( 8-) A 2 CA -42.1 -34.73 33.92 Wrong hand 88 ILE ( 11-) A 2 CA -43.0 -33.43 33.24 Wrong hand 88 ILE ( 11-) A 2 CB -50.2 -34.93 32.30 Wrong hand 96 GLN ( 19-) A 2 CA -35.8 -34.86 33.97 Wrong hand 104 SER ( 27-) A 2 CA -37.8 -36.03 34.32 Wrong hand 107 GLU ( 30-) A 2 C -11.1 -16.11 -0.05 124 GLU ( 47-) A 2 CA -41.9 -35.08 33.98 Wrong hand 149 ILE ( 72-) A 2 CB -49.5 -34.00 32.30 Wrong hand |
The whole structure 1ACP is not very good. take a look at the Ramachandran plot of the second model (that is the one from which I took the two isoleucines):
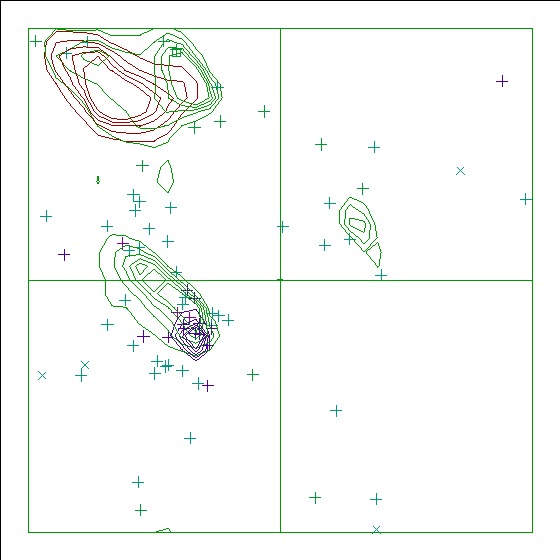
|
Figure 36. Ramachandran plot for the second model of 1ACP. |
Later, the same molecule was solved many times by X-ray. One example is 1L0H. Below, you can see how different a 12 year younger X-ray structure can be...
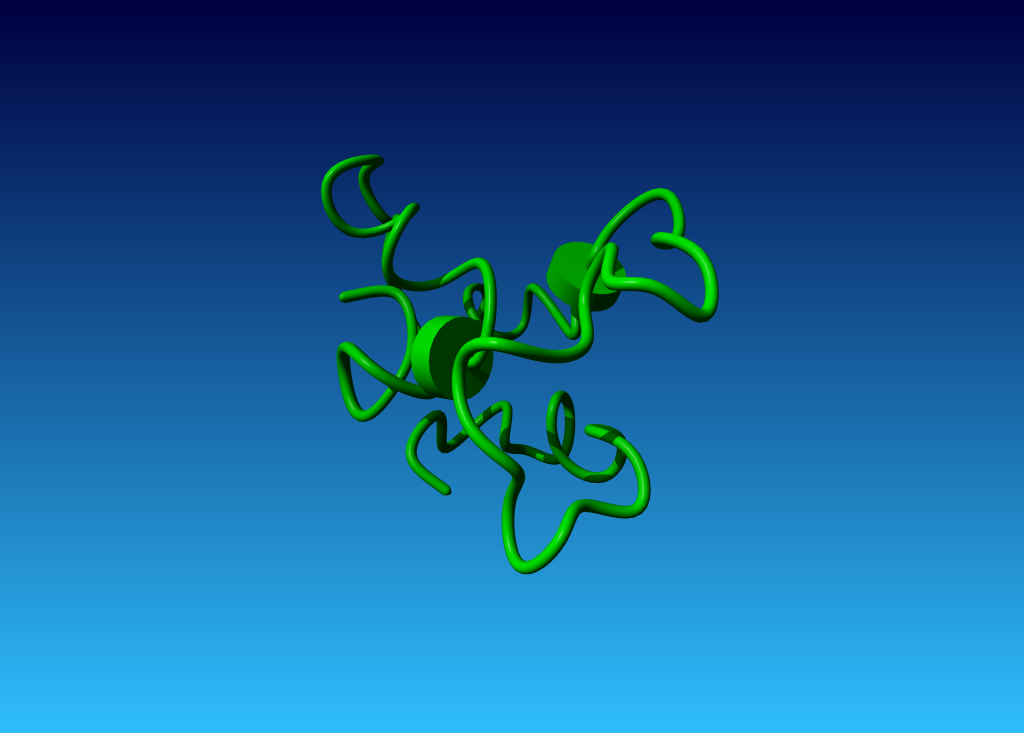
|
Figure 37. The very poor 1ACP structure. |

|
Figure 38. The much better 1L0H structure. |