In this chapter I will discuss many of the messages you can get in section 1 of the validation report. Each paragraph will look like a real error message in a real report, but be aware that I just cut-n-pasted things from a series of reports. Therefore the topics will not be numbered, and sometimes multiple error messages might be internally inconsistent.
The space group name given represents a non-standard setting.
Space group name: P 1 1 21
Conventional space group : P 1 21 1
The space group symbol indicates a different class than the unit cell given on the CRYST1 card of the PDB file.
Possible cause: The unit cell may have pseudo-symmetry, or one of the cell dimensions or the space group might be given incorrectly.
Crystal class of the cell: HEXAGONAL
Crystal class of the space group: MONOCLINIC
Space group name: P 1 21 1
The unit cell in the CRYST1 card of the PDB file contains vectors with lengths smaller than 1.54 Angstrom.
Possible cause: Probably one or more of the values is mistyped, or the CRYST1 card does not conform to the FORMAT given in the PDB specification.
The CRYST1 cell dimensions
A = 1.000 B = 1.000 C = 70.800
Alpha= 90.000 Beta= 90.000 Gamma= 90.000
|
By the way, if you look in the PDB file you see that this is the result of X-ray fiber diffraction. Scientifically it is thus not wrong, but administratively highly inconvenient.
The unit cell in the CRYST1 card of the PDB file contains vectors with lengths smaller than 1.54 Angstrom.
Possible cause: Probably one or more of the values is mistyped, or the CRYST1 card does not conform to the FORMAT given in the PDB specification.
The CRYST1 cell dimensions
A = 0.000 B = 0.000 C = 0.000
Alpha= 90.000 Beta= 90.000 Gamma= 90.000
|
And this one is an NMR file. So, again administratively highly inconvenient because the convention is that the 'cell dimensions' of an NMR structure are (1.0,1.0,1.0,90.0,90.0,90.0).
The space group symbol on the CRYST1 card of the PDB file is B 2 (monoclinic), which is a non-standard c-unique setting for C 2.
The CRYST1 cell dimensions
A = 185.500 B = 72.000 C = 73.000
Alpha= 90.000 Beta= 90.000 Gamma= 77.700
|
The crystallographic unit cell does not conform to the convention that all non-orthogonal angles in a non-triclinic cell should be obtuse.
The CRYST1 cell dimensions
A = 185.500 B = 72.000 C = 73.000
Alpha= 90.000 Beta= 90.000 Gamma= 77.700
|
The crystallographic unit cell does not conform to the convention that a triclinic cell should be specified as having either three obtuse (type II) or three acute angles (type I).
The CRYST1 cell dimensions
A = 32.100 B = 46.200 C = 25.400
Alpha= 105.900 Beta= 113.300 Gamma= 69.700
|
The space group symbol on the CRYST1 card of the PDB file is not one of the valid space groups known to WHAT IF.
Space group name: C 1 21 1
The unit cell in the CRYST1 card of the PDB file contains angles outside the range 60-120 degrees. A conventional cell has angles within these limits.
The CRYST1 cell dimensions
A = 39.821 B = 40.160 C = 39.460
Alpha= 74.420 Beta= 76.440 Gamma= 59.900
|
Space group name: P 1
The crystal class of the conventional cell is different from the crystal class of the cell given on the CRYST1 card. If the new class is supported by the coordinates this is an indication of a wrong space group assignment.
The CRYST1 cell dimensions
A = 32.100 B = 46.200 C = 25.400
Alpha= 105.900 Beta= 113.300 Gamma= 69.700
Dimensions of a reduced cell
A = 25.400 B = 32.100 C = 46.200
Alpha= 69.700 Beta= 74.100 Gamma= 66.700
Dimensions of the conventional cell
A = 58.964 B = 25.400 C = 46.215
Alpha= 90.050 Beta= 112.226 Gamma= 90.006
Transformation to conventional cell
2.000000 0.000000 1.000000
0.000000 0.000000 -1.000000
-1.000000 1.000000 0.000000
|
Crystal class of the cell: TRICLINIC
Crystal class of the conventional CELL: MONOCLINIC
Space group name: P 1
Bravais type of conventional cell is: C
The plot shows the RMS differences between two similar chains on a residue-by-residue basis. Individual "spikes" can be indicative of interesting or wrong residues. If all residues show a high RMS value, the structure could be incorrectly refined.
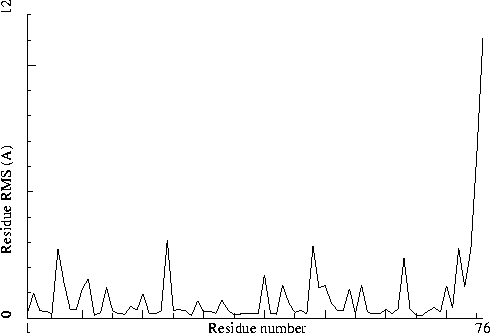
|
Figure 40. The NCS plot in section 1. Labeled with: |
Note: Non crystallographic symmetry backbone difference plot The plot shows the differences in backbone torsion angles between two similar chains on a residue-by-residue basis. Individual 'spikes' can be indicative of interesting or wrong residues. If all residues show high differences, the structure could be incorrectly refined.
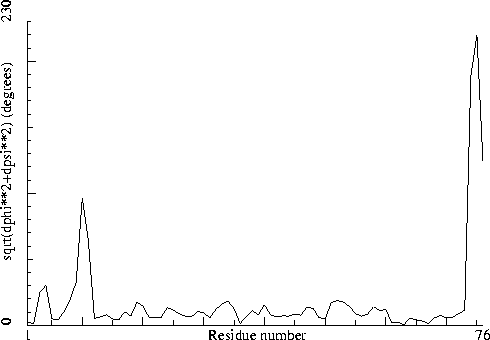
|
Figure 41. The backbone NCS difference plot in section 1. Labeled with: |
In the conventional cell, independent molecules in the asymmetric unit seemingly become symmetry relatives. This fact needs manual checking.