|
After completing the "Amino Acids" part of the Structure section you will: |

|
Figure 52. Load the file HYPOTHASE.pdb in yasara. This is obviously not a real protein, but something we made up to make it easy for you to get used to looking at protein structures. |
EU name: 3D-202
(From: ../step5 )
(Date: Aug 24 2016 ../step)
Question 57:
Find in the structure one of each:
A hydrophobic residue
A hydrophilic residue
A proline
A glutamic acid
A helix
A strand
A sheet
A core residue
A surface residue
A strand with alternating hydrophobic and hydrophilic residues
Answer
Question 58:
1) Study the residues that form the one single helix in
this molecule.
2) Which residues does YASARA think belong to this helix?
3) And what do you think? (Hint, look along the axis of the helix; look at the
direction of C=O groups, look at the hydrogen bonds, etc).
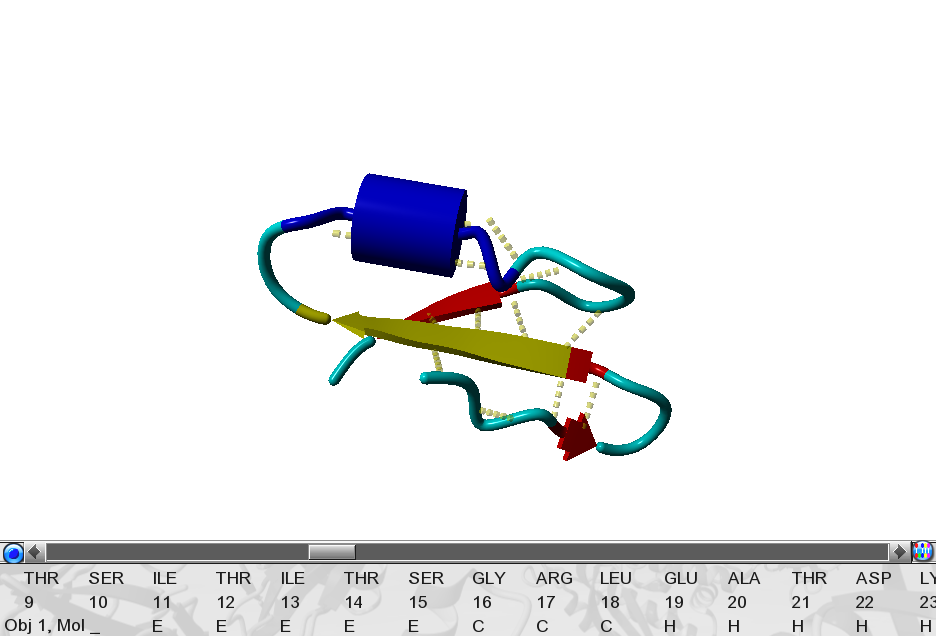
|
Figure 53. Demonstration of YASARA visualization options. |
Question 59:
Try to reconstruct this figure at your screen.
Do not continue before the assistant has checked that the figures match exactly.
(And in case the assistant is not immediately available, look in the File menu
how you can save the situation as a Scene...).