Step 1. Find BLAST. It is part of the MRS bioinformatics suite. And MRS can be found in the 'Pointers' window.
The blue arrows tell you where to click...

|
Step 1. Find BLAST. It is part of the MRS bioinformatics suite. And MRS can be found in the 'Pointers' window. |

|
After a second or so, BLAST is done. Now click on line that holds your results (you most likely will have only one line), and you get the next picture. |
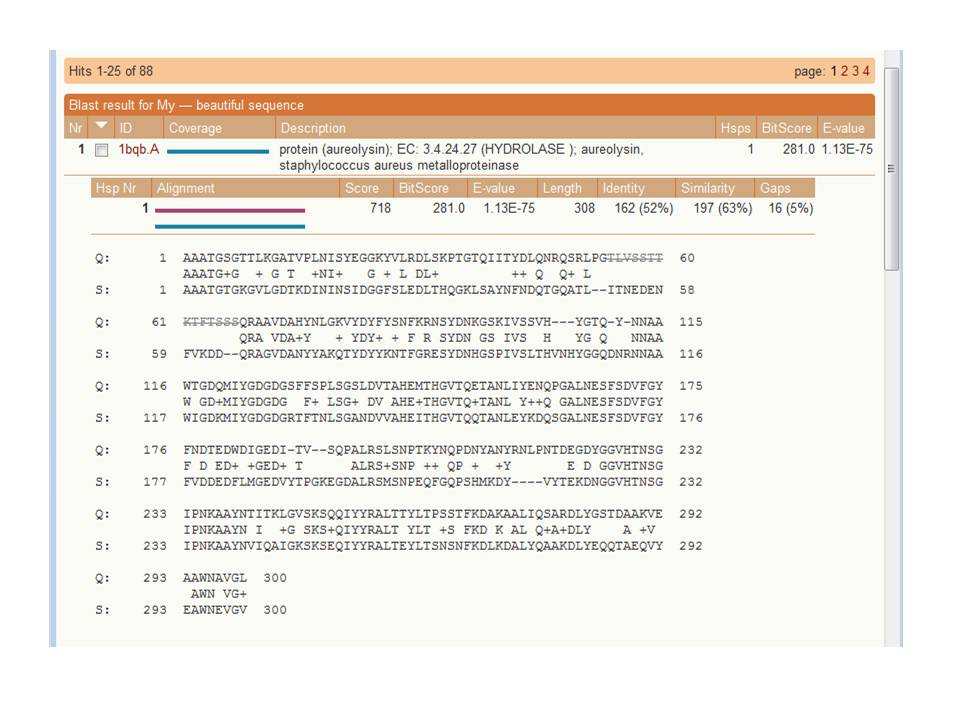
|
The alignment made by BLAST. Feel free to ask the assistants what all
things mean, but this is not exam-material... |