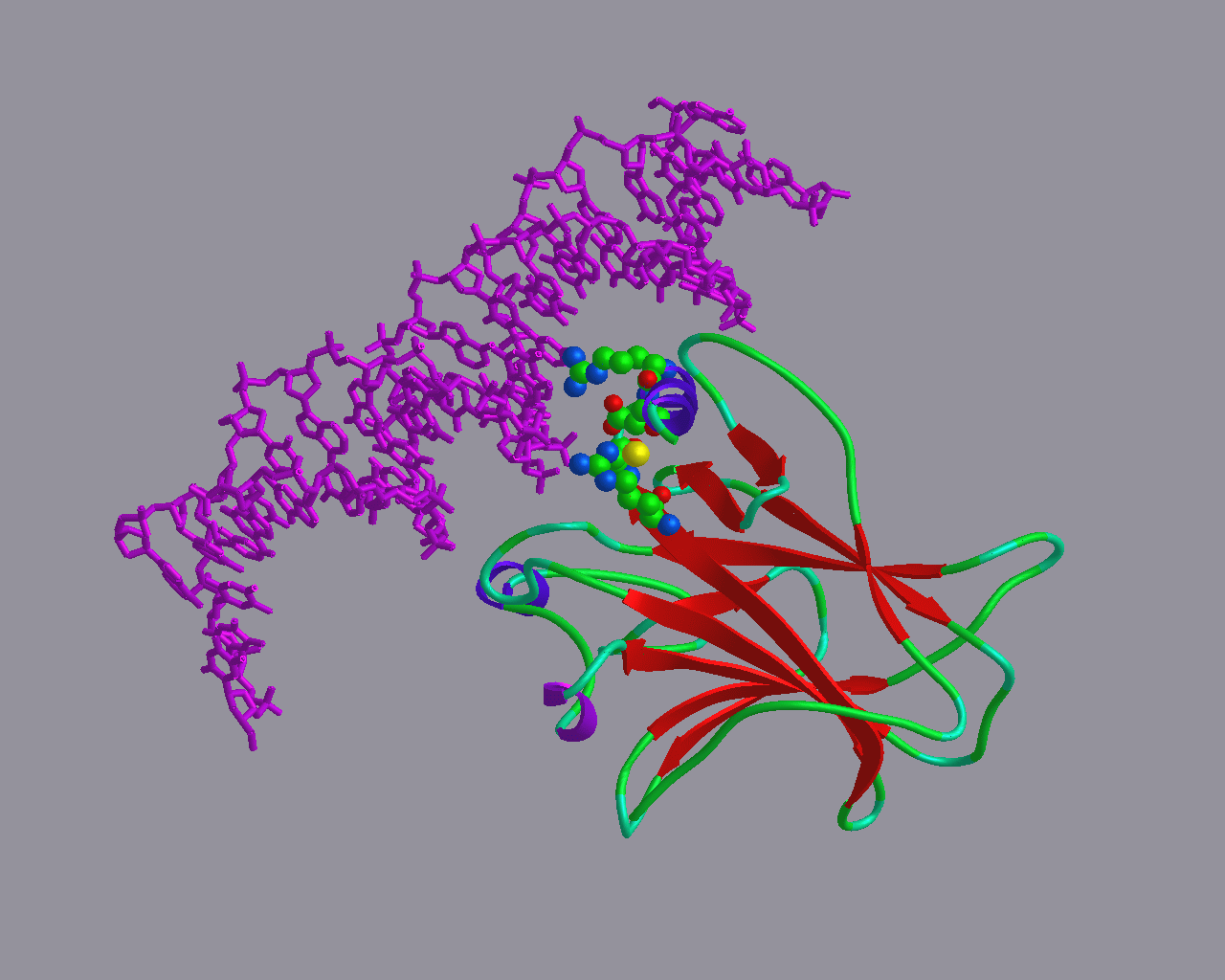
The model of P63 immediately made clear that the mutations that cause the EEC syndrome (I will save you what EEC stands for, I cannot pronounce it anyway): Disruption of the DNA-binding capability.
We often do little collaborative projects with biomedical scientists. Besides helping our colleagues, we do these projects mainly for two reasons:
This page lists a series of these projects. This list is far from complete, but, you know, there are only 24 hours in a day. You can find dozens more projects at Hanka's web pages.
P63 is a transcription regulator. The group of Han Brunner discovered that certain inheritable mutations in this molecule cause very unpleasant diseases. Their obvious second question was why these mutants were not simple, insignificant SNPs like the thousands of other differences between you and me, and what are the molecular reasons for causing the disease.

|
The model of P63 immediately made clear that the mutations that cause the EEC syndrome (I will save you what EEC stands for, I cannot pronounce it anyway): Disruption of the DNA-binding capability. |
Heterozygous Germline mutations in the p53 homolog p63 are the cause of
EEC syndrome.
J.Celli, P.Duijf, B.C.J.Hamel, M.Bamshad, B.Kramer, A.P.T.Smits,
R.Newbury-Ecob, R.C.M.Hennekam,
G.v.Buggenhout, A.v.Haeringen, C.G.Woods, A.J.v.Essen, R.d.Waal,
G.Vriend, D.A.Haber,
A.Yang, F.McKeon, H.G.Brunner, H.v.Bokhoven
Cell (1999) 99, 1-20.
The group of Ida vd Klei is interested in alcohol oxidase. She came to us with the simple question: "How does this molecule multimerise, and what is the role of multimerisation in its function"? We built a model, and started looking at it. But there was nothing special about anything to be found in this model. Fortunately, we have a large series of visualization techniques available that are implemented in programs like WHAT IF or YASARA to help us do just that, look at protein structures and analyze them.
This allowed us to suggest that Ton Visser uses his spectroscopic magic to count the tryptophans involved in each of the steps. This answered the question.
So, in this case, bioinformatics did not solve the problem, but at least it revealed how it could be solved experimentally. And even though it hurts my theoretician's heart to say it, but the Dutch saying "meten is weten" (to measure is to know) is true.....
A scientists from Texas one day asked me how a certain protein fragment caused dimerisation. That protein fragment is:
MELLSPPLRDIDLTGPDGSLCSFETADDFYDDPCFDSPDLRFFEDLDPRL | | | | | || || | | || |
It struck me that the protein had very many negative charges (indicated by vertical bars). As he told me that this protein had something to do with transcription regulation (which is mainly done by proteins with helical motifs), I built the structure of this fragment as one big helix.
Octamer motif proteins are transcription factors that function in the embryo. A long time ago, I modelled Oct 4 for Hans Scholler. This protein consists of two domains that each sit at the opposite site of the DNA. These two domains look like each other. Later it was found out that two of these pseudo dimers bind at the same DNA site. And as the two halves of dimer are connected by a long and flexible linker, we now got stuck with two solutions:
Synergism with the Co-activator OBF-1 (OCA-B, BOB-1) Is Mediated by a
Specific POU Dimer Configuration.
Tomilin A, Remenyi A, Lins K, Bak H, Leidel S, Vriend G, Wilmanns M,
Scholer HR.
Cell. (2000) 103: 853-864.
PubMed Abstract
Sometimes, we collaborate with people who do not have easy access to visualization software. Clearly, those people should look into YASARA , but what can I say. The CMBI supports the JMOL initiative. And that allows us to present molecules to everybody with a computer with a browser. The picture below is of a model of a protein called shifl. And, if you click on "Activate with JMOL" the molecule will become interactive with JMOL.

|
This is a picture of shifl. If you click on "Activate with JMOL" you can look at shifl with JMOL. The JMOL files and pages are 100% produced by WHAT IF |
|
IMAGE/1crn.pdb Activate with JMOL |