|
After completing the "Amino Acids" part you will know for each of the 20 amino acids: |
Amino acids are at the heart of bioinformatics. Evolution of life is mainly governed by individual amino acid changes that slowly accumulate over the eons (this is not the whole story, of course, but surely a large part of it). If we want to align two sequences, we align amino acids. And your body's workhorses, the enzymes, consist of mainly amino acids, and if we want to modify a protein using protein engineering techniques, we exchange amino acids.
Therefore, a thorough understanding of the amino acids is needed. At the end
of this course you will all know the twenty amino acids as if they are your
friends. You will know their names, abbreviations, physico-chemical
characteristics, and a whole lot more. At several moments we will relate things
to thermodynamics and we will mention the "rule of 10":
![]()
The most important characteristic of amino acids is their hydrophobicity,
but their size and charge are also important. The hydrophobicity is intimately
connected to the role of water:
![]()
The table below lists 5 items. You should now briefly look at this material, and study it either at your leisure at home, or when you need it while going through the questions later today or in the coming weeks. The five items are:
These small amino acid pictures will start rotating if you click them.
 Alanine
Alanine
 Cysteine
Cysteine
 Aspartic acid
Aspartic acid
 Glutamic acid
Glutamic acid
 Phenylalanine
Phenylalanine
 Glycine
Glycine
 Histidine
Histidine
 Isoleucine
Isoleucine
 Lysine
Lysine
 Leucine
Leucine
 Methionine
Methionine
 Asparagine
Asparagine
 Proline
Proline
 Glutamine
Glutamine
 Arginine
Arginine
 Serine
Serine
 Threonine
Threonine
 Valine
Valine
 Tryptophan
Tryptophan
 Tyrosine
Tyrosine
Question 33:
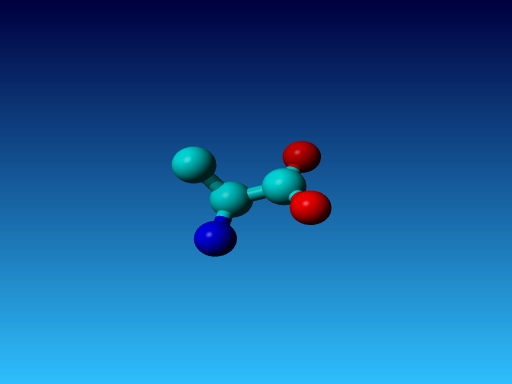
|
Figure 41. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 34:
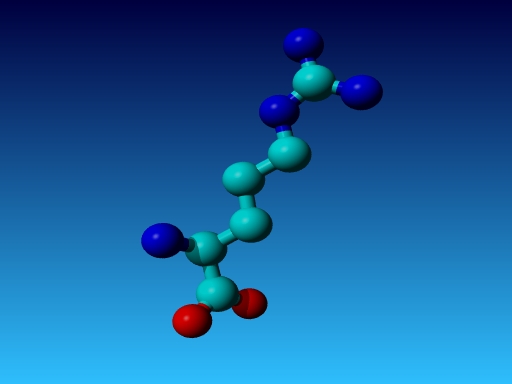
|
Figure 42. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 35:
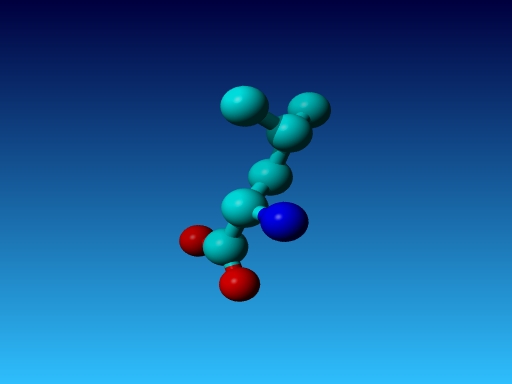
|
Figure 43. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 36:
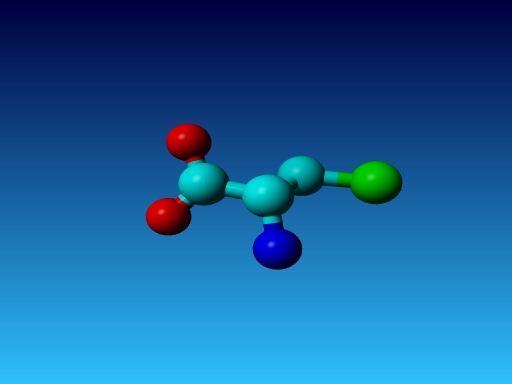
|
Figure 44. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 37:
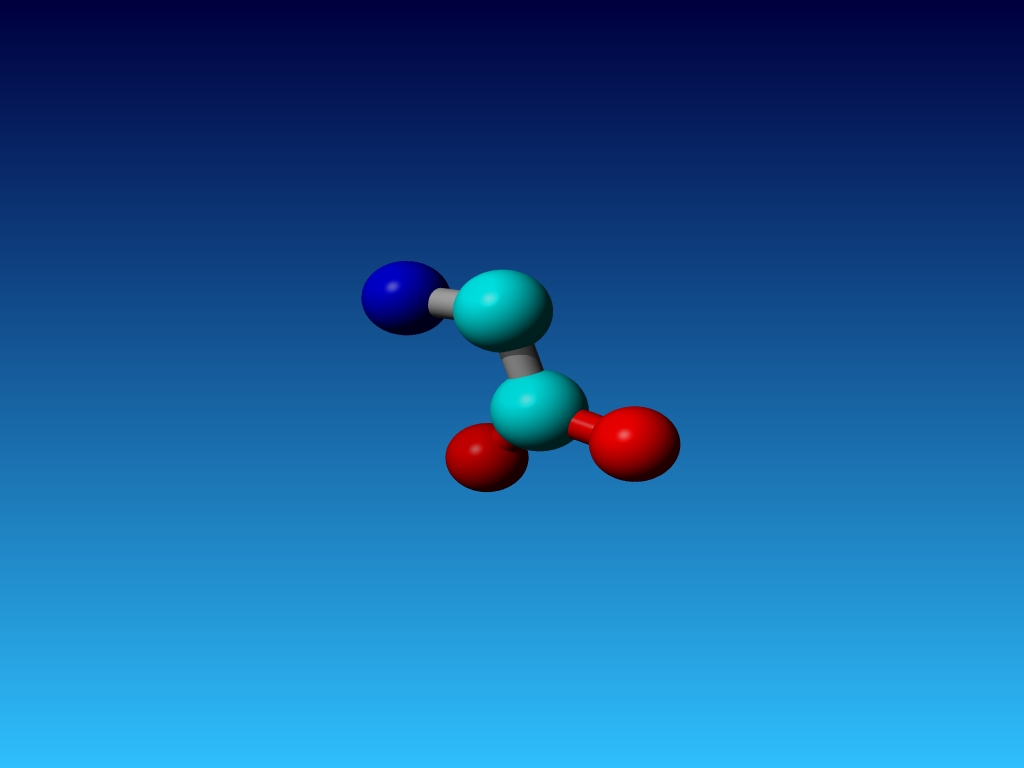
|
Figure 45. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 38:
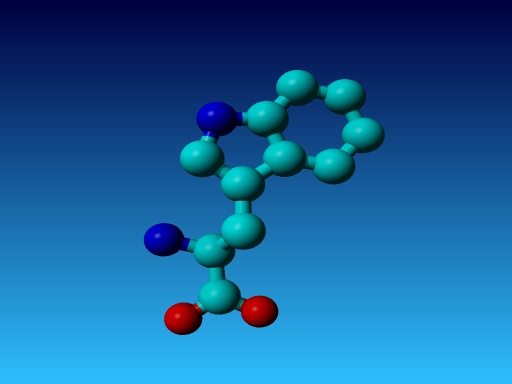
|
Figure 46. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 39:
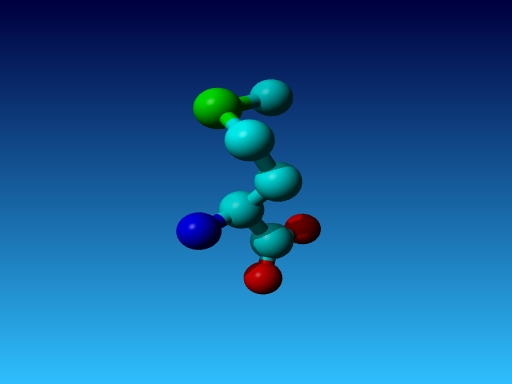
|
Figure 47. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 40:
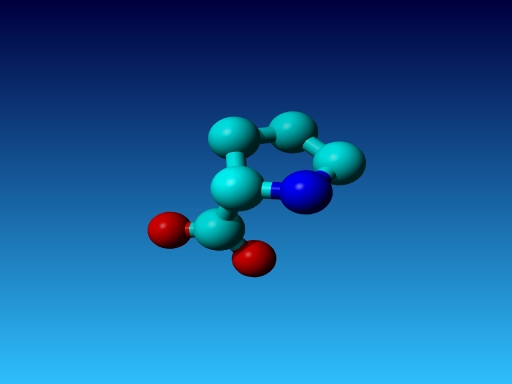
|
Figure 48. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 41:
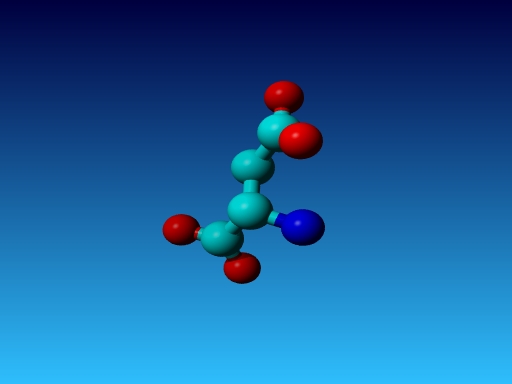
|
Figure 49. Which residue is this, and what do you know about its special characteristics? |
Answer
Question 42:
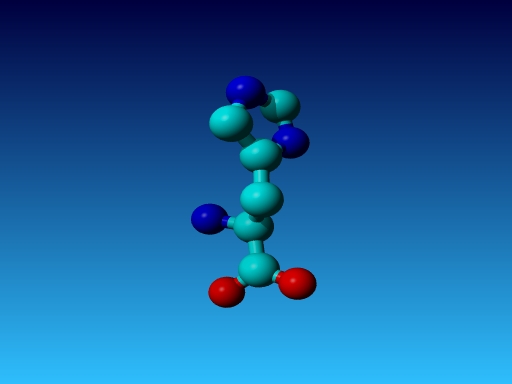
|
Figure 50. Which residue is this, and what do you know about its special characteristics? |
Answer
At this moment you should know your amino acids. before continuing the course, do the amino acid quiz.
Test yourself.
Question 43: Below you see all but the last quiz-question repeated in almost the same words as used in the quiz. You don′t have to write down the answers because the quiz-system tells you whether your answers were correct or wrong.
Answer
Supplemental material
You can use at a very rough rule-of-thumb that the hydrophilicity of a side chain is determined by the number of N and O atoms in the side chain. The exception is the N in tryptophan that is almost entirely hydrophobic. Charged N and O atoms are slightly more hydrophilic than non-charged ones. And given an equal number of N and O atoms the one with the most carbons or sulphurs is the more hydrophobic one. Obviously, this rule-of-thumb is very crude. And further down this page we will see that it is probably impossible to even make a good rule for hydrophobicity determination of amino acids. However, as hydrophobicity certainly is the most important characteristic of the amino acid, everything that helps you work with in in your head helps you become a better bioinformatician. |
Question 44: Use the rule-of-thumb that is boxed above this question (on the website not in this handout...), and look at the sheet with amino acid structure formulas to find 10 pairs of amino acids that look very much like each other, actually they should look so much like each other that they differ by no more than one atom. You can start with Ala-Ser and Thr-Ser, but the rest you have to find yourself. In each case write the more hydrophobic residue of the pair to the left.
Answer
Question 45:
The scorings matrix used by BLAST reflects how much residues look like each other.
If you had to design a scorings-matrix for BLAST, would you give the pairs you
listed in the previous question hsigh points or low points?
And where would you put the highest points in your matrix?
Obviously, the rule-of-thumb is just that, a rule-of-thumb. In reality you should determine the pKa of each titratable group, take into account the pH, the salt concentration (and the type(s) of salt involved), the temperature, etcetera. But for now, the rule-of-thumb is good enough.
Question 46:
Given the rules of thumbs, and your pairwise sorting, make a list of
all amino acids and sort them by hydrophobicity. Start with Trp as the most
hydrophobic one at the far left, and end with Arg as the most hydrophilic
one at the far right.
Discuss the answer with an assistant before continuing with the
subsequent questions.
Question 47:
Look up the pKa values of the titratable groups (
http://swift.cmbi.umcn.nl/teach/aainfo/index.shtml; section 3.3).
Why are the pKa values given for only 7 amino acids?
What are the pKa values of the two positive residues? And of the two
negative values? And what is the pH in the cytosol of a human cell? And why
are these questions combined in one question; in other words what is the
deeper meaning here?
The pKa of Cys is ~9.3. Does that make Cys a basic residue?
Now go back to the hydrophobicity scales and think about what the
assistants told you about the position of Lys. Does that story now
make sense?
You are not the only ones who have made a hydrophobicity scale. Others have done that many years ago already. And many scales were made with very sophisticated techniques.
Look up the hydrophobicity scales (
http://swift.cmbi.umcn.nl/teach/aainfo/hydro.shtml)
![]() and first find out what this is all about; what is in those scales; how were they
determined; etc.
and first find out what this is all about; what is in those scales; how were they
determined; etc.
Question 48:
Look at the scales mentioned above (on the website, not in the handout) and answer:
Which is the most hydrophobic residue?
What is wrong with the previous question?
What is the difference between scales 1&4
versus 2&3? In other words,
which experiments were done to arrive at the values in these scales?
Explain the different position for cysteine in 1&4
versus 2&3.
Explain the position of proline in the scales. Is it located where you
expected it?
Explain the positions of arginine en lysine in the different scales.
Can you think of a possible reason that arginine and lysine are hardly ever
found inside a protein, while glutamic acid and aspartic acid can
occasionally be found there?
There is, of course, a strong relation between hydrophobicity and the preference of amino acids for the surface or the inside of the protein. But this is a preference, and not a law. Hydrophobic residues can be found at the surface and in the core of the protein, whereas hydrophilic ones tend to be found much more often at the surface than inside the protein.
You often see in standard text books that hydrophobic residues sit in the core, and hydrophilic ones at the surface of proteins. In the next step of this course you can see for yourself that that is not true. |
Look up the page about solvent accessible surfaces
(
http://swift.cmbi.umcn.nl/teach/aainfo/access.shtml)
Note: One water molecule has a diameter of about 2.8 Ångström and it can
touch a surface of about 10 Ångström2 of the surface of a protein.
A SEA of 30 Ångström2 means that about three water molecules can
touch this amino acid (on average in the dataset used by the
bioinformaticians who produced this table).
Question 49:
Look up the above scales (on the website, not in the handout) and
answer the (simple) questions:
Which amino acids will be more accessible to solvent (water), those at the
exterior (outside) or those in the interior (inside) of the protein?
Describe in your own words the meaning of the numbers in the table.
(Just a small side-step: How do you think these numbers are being calculated?
In other words, what does the computer algorithm look like)?
Which amino acids are on average most accessible?
Which amino acid is on average least accessible?
What is so dangerous about questions like the previous two?
Try to explain why methionine is relatively often found at the outside of
the protein.
Proline has an unexpected SEA distribution. Can you explain why?
When thinking about residue hydrophobicity and the surface location of proteins there is a nice, for many people surprising, realisation.
Question 50:
If you observed a polar residue in the core of a protein, and you mutate it into
a hydrophobic residue, the protein often gets more stable. But when you observe
a hydrophobic residue at the surface and mutate it into a polar one, you
normally don't see any effect on the stability.
Think about the entropy of water. Realize that stability is measured
as the temperature at which the protein folding process (folded --
unfolded is just in equilibrium.
And now the question: Why is this?
Question 51:
Draw the general structure of one amino acid (use R to indicate the side chain).
Indicate the N-terminal side and the C-terminal side of the amino acid. Also put the names of
the backbone atoms in the drawing (N, Cα, C, O).
Draw a dipeptide Val-Lys, (that is two amino acids joined by a peptide bond)..
Draw the dipeptide Lys-Val.
Are those dipeptides identical?
Answer
Question 52:
Which atoms in the protein backbone can form hydrogen bonds?.
Which backbone atom donates the hydrogen atom (hydrogen bond donor)?.
Which backbone atom accepts the hydrogen atom (hydrogen bond acceptor)?.
And, actually, now that we are talking about it: What is a hydrogen bond?.
Answer
Question 53: The main reason for our study of the amino acids is understanding them well enough to interpret (and if needed correct) alignments. So, lets take a look at some alignments. It should be possible to answer these questions using no more than a bit of brainpower (to think about the relative importance of residues) and the information provided in the amino acid information pages. For each pair of alignments listed below, determine which is the best one (left or right) and explain why.
1 LLLWASLLL LLLWASLLL LLLLWSLLL LLLWSLLLL |
2 LLLCANLLL LLLCANLLL LLLLCNLLL LLLCNLLLL |
3 LLLRAGLLL LLLRAGLLL LLLLRGLLL LLLRGLLLL |
4 LLLHANLLL LLLHANLLL LLLLHNLLL LLLHNLLLL |
5 LLLPAKLLL LLLPAKLLL LLLLPKLLL LLLPKLLLL |
Answer
Question 54:
For each amino acid, list its 1-letter code, its 3-letter code, its full name,
whether it is hydrophobic/hydrophilic/inbetween, its size (small/medium/large),
its charge at pH 6 a 7, its secondary structure, and anything special you know
about this amino acid.
Feel free to ask the assistants for an empty amino acid sheet to make this
exercise administratively easier. (There is an extra copy for you to print
under the heading 'test exams').
Question 55: Go through the amino acid seminar and figure out what are special characteristics of the amino acids. Try to summarise these characteristics in 1 word per characteristic otherwise it won't fit in the small box.
Answer|
|