
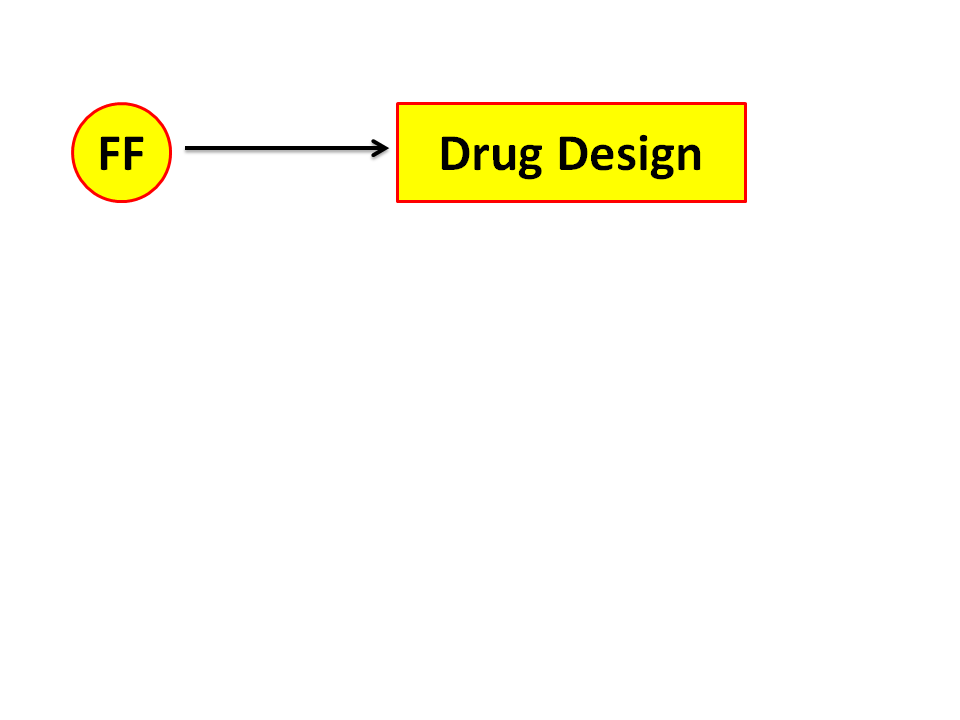
Figure 3. We start this summary picture series with Drug Design. Drug Design
has drug docking as a big sub-topic. Drug design and drug docking are the
largest fields in terms of employing chemists.
![]()
|
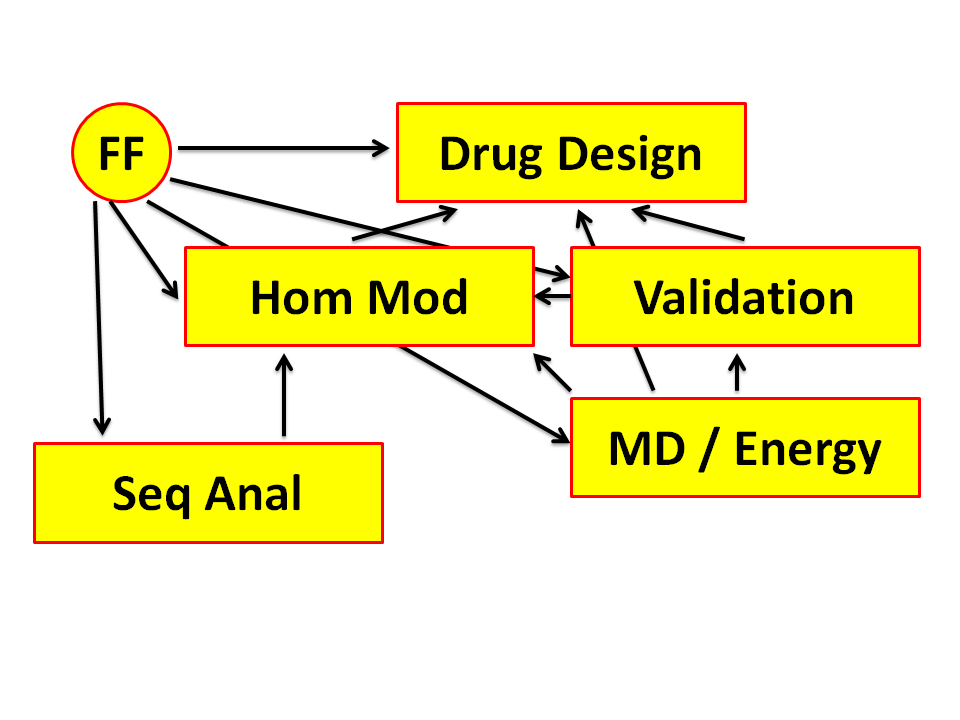
The course will revolve around six topics that all relate to each other: Drug Design and Drug Docking; Force Fields; Homology Modelling; Structure Validation; Molecular Dynamics; Sequence Alignment. |
The seminars all depend a bit on each other. That makes it difficult to choose an order. It also means that we will often refer forward ("In the next seminar you will hear that...").
After the six regular seminars there will be one long session in which all six seminars will be briefly repeated.

|
Figure 3. We start this summary picture series with Drug Design. Drug Design
has drug docking as a big sub-topic. Drug design and drug docking are the
largest fields in terms of employing chemists.
|
|
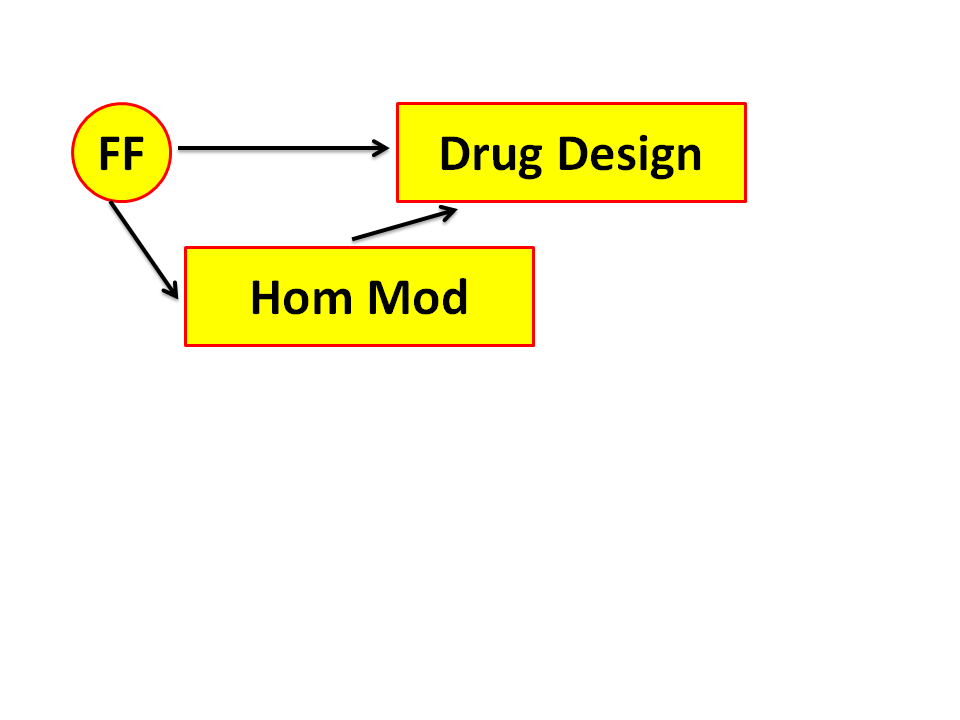
Figure 4. Drug design makes extensive use of Force Fields. Several of those
Force Fields will be discussed in this course.
|
|
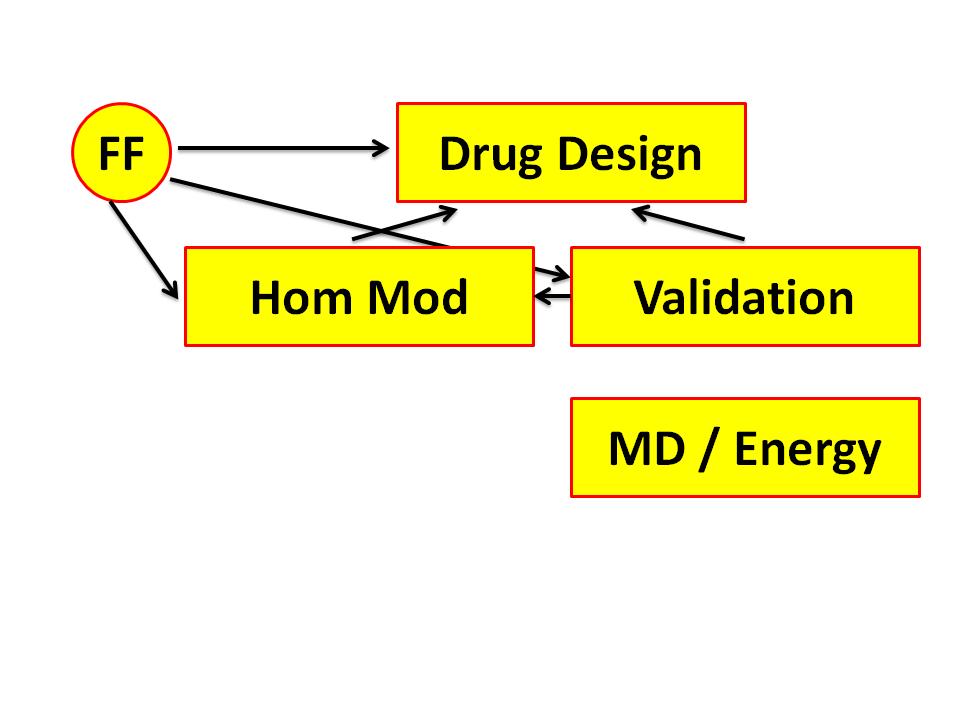
Figure 5. Virtual screening is the term used for testing for a large number
of small molecules if they can be docked in a pocket in a target. These
targets normally are proteins. If the structure of the protein has not been
solved yet by either Xray crystallography or NMR spectroscopy, then
we need to predict it. This is best done with Homology Modelling. Homology
Modelling makes extensive use of Force Fields, too.
|
|
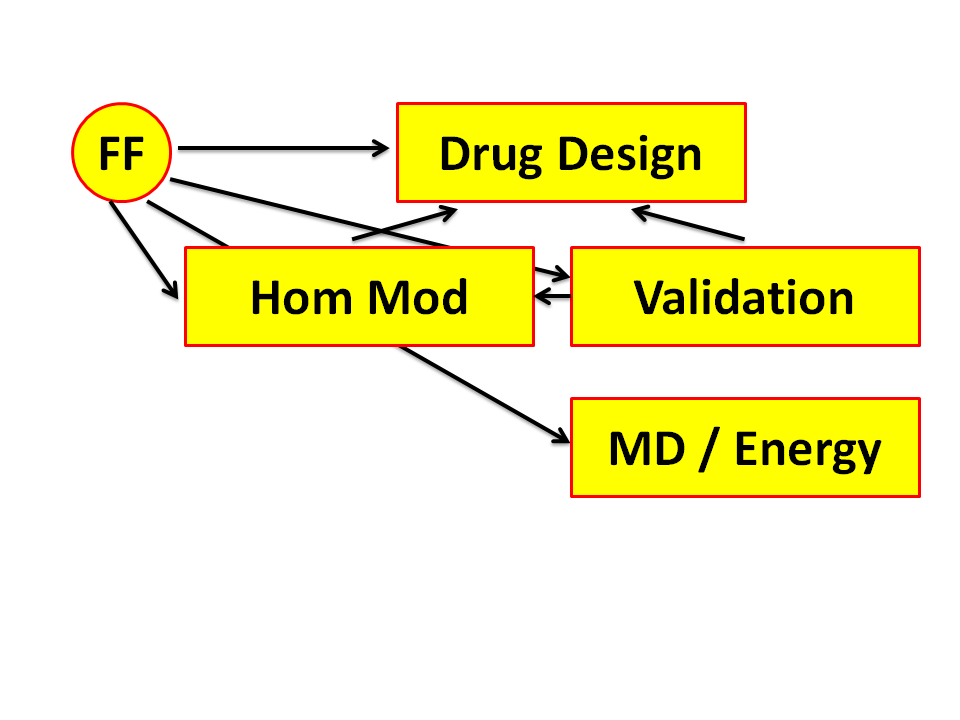
Figure 7. Proteins are not hard as rocks but actually rather flexible. So,
if you see a protein structure, you see one possible structure that
accidentally could be crystallized. If your drug seems not to fit in this
conformation, then you might discard it as a lead for drug design. But
if another conformation would be ideal for this drug, then you actually discard
a potentially very good opportunity. This is where Molecular Dynamics can help.
With Molecular Dynamics you can predict how a protein moves around in its
conformational space.
|
|
Figure 8. Homology Modelling uses a rather extensive Force Field, and we will spend an entire seminar to explain its intricacies. |
|
Figure 10. Homology Modelling requires that the homology between your protein
of interest and the homologous protein with know structure gets worked out at
the level of the individual amino acids. This is called sequence alignment .
Sequence alignments, though, can provide much more information and a few
examples will be presented in the Sequence Alignment seminar.
|